Consider the following model:
This is known as a change-point model. The parameter is called the change-point. is the indicator function which takes the value 1 if and 0 otherwise. The function equals for times and equals for times . Therefore this model states that the level of the time series equals until a time at which point it switches to . The value of is therefore called the changepoint. From the given data , we need to infer the parameter as well as . The unknown parameter makes it a nonlinear model. If were known, this will become a linear regression model with -matrix given by
For parameter estimation and inference, we first compute RSS():
and then minimize over to obtain the MLE of . After finding , we can find the MLEs of the other parameters as in linear regression with known .
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm# Here is a simulated dataset having a change point:
n = 10000
mu1 = 0
mu2 = 0.4
dt = np.concatenate([np.repeat(mu1, n/2), np.repeat(mu2, n/2)])
sig = 1
rng = np.random.default_rng(seed = 42)
errorsamples = rng.normal(loc = 0, scale = sig, size = n)
y = dt + errorsamplesplt.figure(figsize = (15, 6))
plt.plot(y)
plt.show()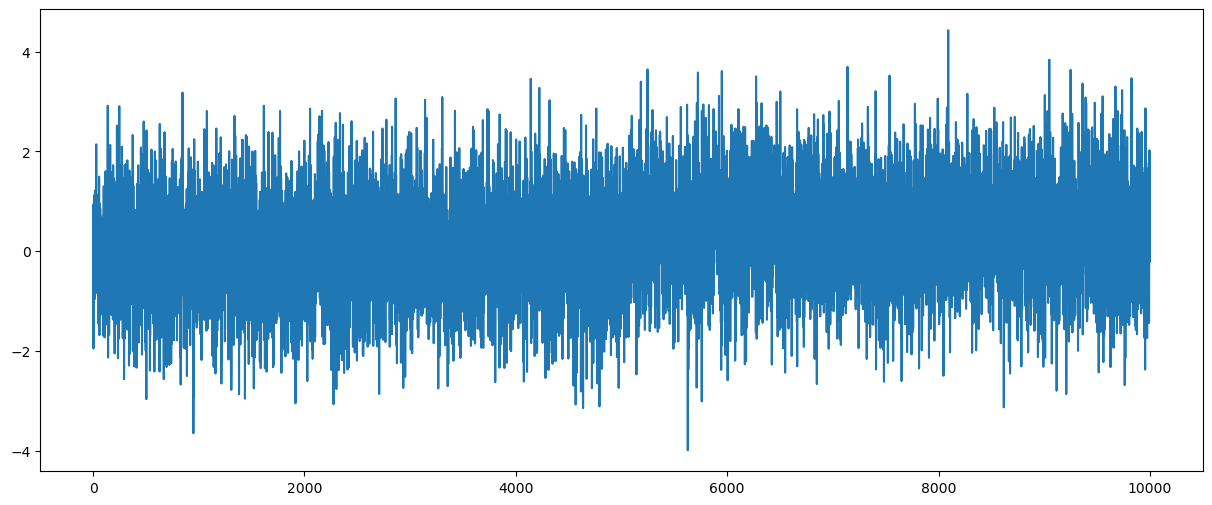
def rss(c):
x = np.arange(1, n + 1)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc])
md = sm.OLS(y, X).fit()
rss = np.sum(md.resid ** 2)
return rssallcvals = np.arange(1, n)
rssvals = np.array([rss(c) for c in allcvals])plt.plot(allcvals, rssvals)
plt.show()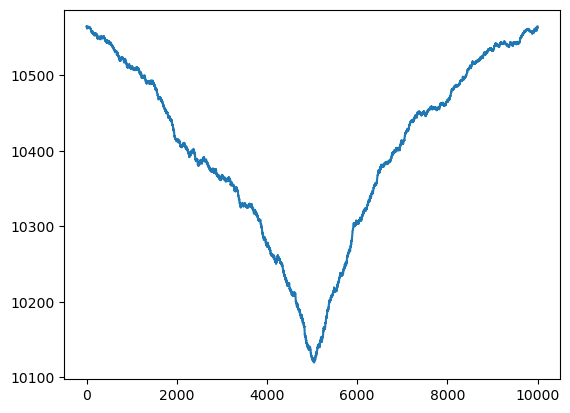
c_hat = allcvals[np.argmin(rssvals)]
print(c_hat)5045
# Estimates of other parameters:
x = np.arange(1, n + 1)
c = c_hat
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc])
md = sm.OLS(y, X).fit()
print(md.params) # this gives estimates of beta_0, beta_1 (compare them to the true values which generated the data)
b0_est = md.params[0]
b1_est = md.params[1]
b0_true = mu1
b1_true = mu2 - mu1
print(np.column_stack((np.array([b0_est, b1_est]), np.array([b0_true, b1_true]))))
rss_chat = np.sum(md.resid ** 2)
sigma_mle = np.sqrt(rss_chat / n)
sigma_unbiased = np.sqrt(rss_chat / (n - 2))
print(np.array([sigma_mle, sigma_unbiased, sig])) #sig is the true value of sigma which generated the data[-0.01930042 0.42189817]
[[-0.01930042 0. ]
[ 0.42189817 0.4 ]]
[1.00596521 1.00606582 1. ]
# Plotting the fitted values:
plt.plot(y)
plt.plot(md.fittedvalues, color = 'red')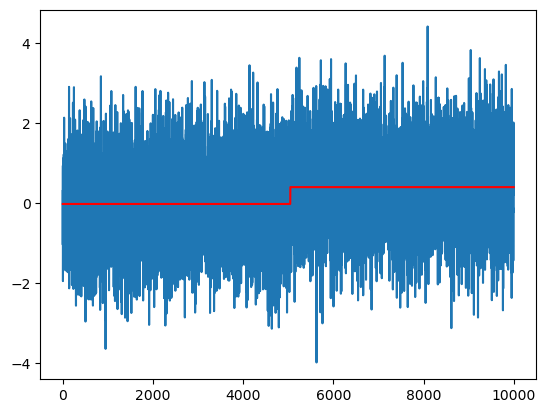
The Bayesian posterior for is:
where and denotes the determinant of .
As before, it is better to compute the logarithm of the posterior (as opposed to the posterior directly) because of numerical issues.
def logpost(c):
x = np.arange(1, n + 1)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc])
p = X.shape[1]
md = sm.OLS(y, X).fit()
rss = np.sum(md.resid ** 2)
sgn, log_det = np.linalg.slogdet(np.dot(X.T, X))
# sgn gives the sign of the determinant (in our case, this should 1)
# log_det gives the logarithm of the absolute value of the determinant
logval = ((p - n) / 2) * np.log(rss) - 0.5 * log_det
return logvalallcvals = np.arange(1, n)
logpostvals = np.array([logpost(c) for c in allcvals])
plt.plot(allcvals, logpostvals)
plt.xlabel('Changepoint')
plt.ylabel('Value')
plt.title('Logarithm of (unnormalized) posterior density')
plt.show()
# this plot looks similar to the RSS plot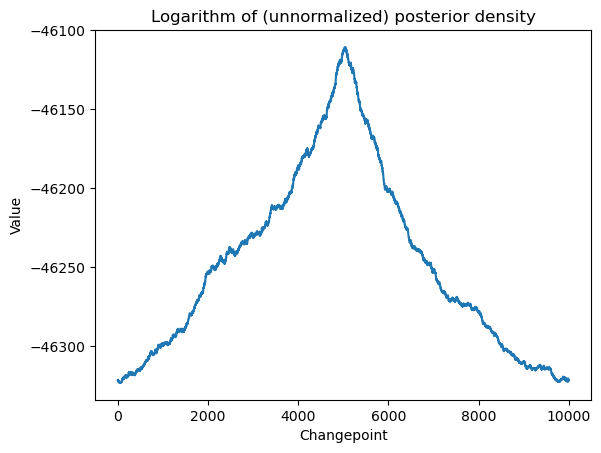
Let us exponentiate the log-posterior values to get the posterior.
postvals_unnormalized = np.exp(logpostvals - np.max(logpostvals))
postvals = postvals_unnormalized / (np.sum(postvals_unnormalized))
plt.plot(allcvals, postvals)
plt.xlabel('Changepoint')
plt.ylabel('Probability')
plt.title('Posterior distribution of change points')
plt.show()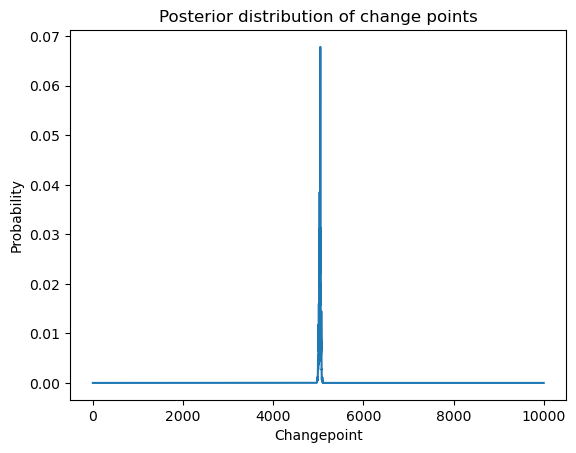
The following code generates posterior samples.
N = 2000
cpostsamples = rng.choice(allcvals, N, replace = True, p = postvals)
post_samples = np.zeros(shape = (N, 4))
post_samples[:,0] = cpostsamples
for i in range(N):
c = cpostsamples[i]
x = np.arange(1, n + 1)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc])
p = X.shape[1]
md_c = sm.OLS(y, X).fit()
chirv = rng.chisquare(df = n - p)
sig_sample = np.sqrt(np.sum(md_c.resid ** 2) / chirv) #posterior sample from sigma
post_samples[i, (p + 1)] = sig_sample
covmat = (sig_sample ** 2) * np.linalg.inv(np.dot(X.T, X))
beta_sample = rng.multivariate_normal(mean = md_c.params, cov = covmat, size = 1)
post_samples[i, 1:(p + 1)] = beta_sample
print(post_samples)[[ 5.05000000e+03 -1.28000430e-02 4.20797852e-01 1.00231554e+00]
[ 5.02500000e+03 8.04560220e-03 4.26058253e-01 1.01677879e+00]
[ 5.05300000e+03 7.02856193e-03 4.00933767e-01 1.00499166e+00]
...
[ 5.06700000e+03 -2.11369872e-02 4.34416448e-01 9.87343752e-01]
[ 5.01700000e+03 -3.12764767e-02 4.09694942e-01 9.93151258e-01]
[ 5.00600000e+03 3.34638353e-03 4.09055268e-01 1.00213666e+00]]
x = np.arange(1, n + 1)
plt.figure(figsize = (15, 6))
plt.plot(y)
for i in range(N):
c = cpostsamples[i]
b0 = post_samples[i, 1]
b1 = post_samples[i, 2]
ftdval = b0 + b1 * (x > c).astype(float)
plt.plot(ftdval, color = 'red')
# Plotting only the change-point samples:
plt.figure(figsize = (15, 6))
plt.plot(y)
for i in range(N):
c = cpostsamples[i]
plt.axvline(c, color = 'red')
plt.axvline(5000, color = 'black') # 5000 is the true value of c
plt.show()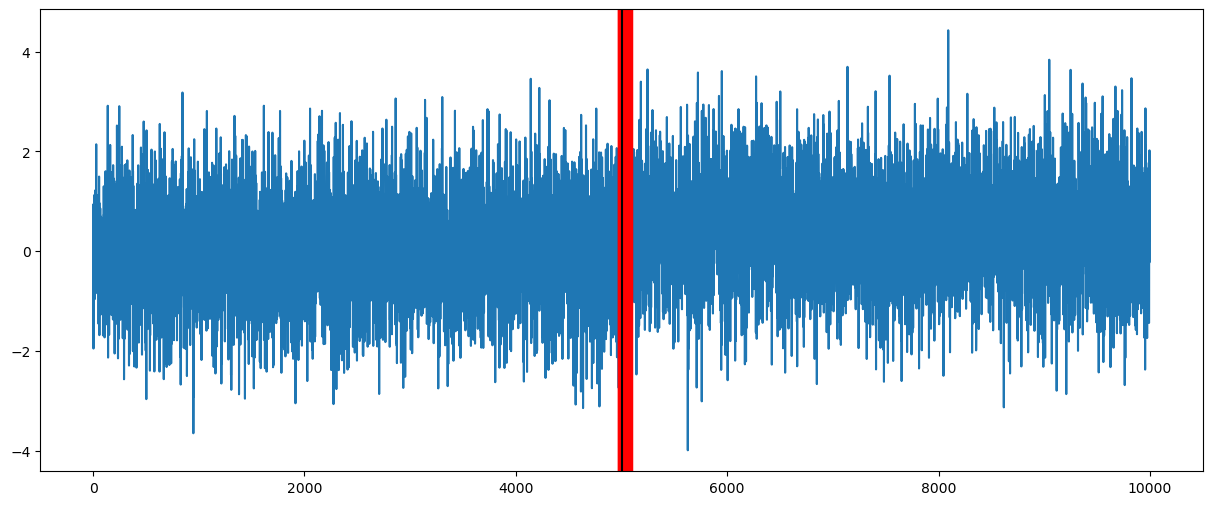
Example with multiple change-points¶
We now consider an example where there are three change points and the model we want to use is:
We work on a simulated dataset that is generated as follows.
sig = 1
mu1 = 0
mu2 = 1.5
mu3 = -1
mu4 = 0
truedt = np.concatenate((np.repeat(mu1, 100), np.repeat(mu2, 100), np.repeat(mu3, 100), np.repeat(mu4, 100)), axis = None)
n = len(truedt)
noise = rng.normal(size = n)
y = truedt + sig * noise
plt.plot(y)
plt.show()
cps_true = np.array([100, 200, 300]) # these are true changepoints
print(cps_true)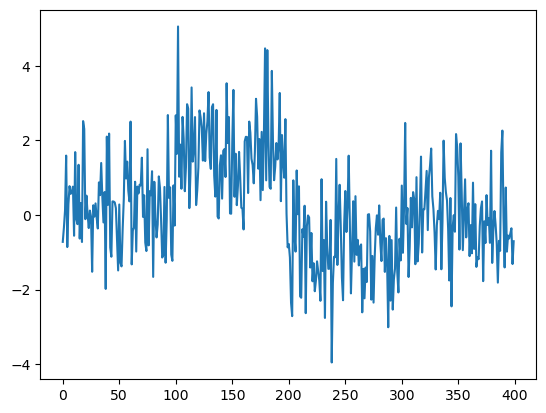
[100 200 300]
The following is the RSS for fitting three change points to the data.
def rss(c):
n = len(y)
x = np.arange(1, n + 1)
X = np.column_stack([np.ones(n)])
if np.isscalar(c):
c = [c]
for j in range(len(c)):
xc = ((x > c[j]).astype(float))
X = np.column_stack([X, xc])
md = sm.OLS(y, X).fit()
ans = np.sum(md.resid ** 2)
return ansc1_gr = np.arange(75, 126)
c2_gr = np.arange(175, 226)
c3_gr = np.arange(275, 326)
X, Y, Z = np.meshgrid(c1_gr, c2_gr, c3_gr)
g = pd.DataFrame({'x': X.flatten(), 'y': Y.flatten(), 'z': Z.flatten()})
g['rss'] = g.apply(lambda row: rss([row['x'], row['y'], row['z']]), axis = 1)
# this is taking about 49 secs to run on my laptopmin_row = g.loc[g['rss'].idxmin()]
print(min_row)
c_opt = np.array(min_row[: -1])x 100.000000
y 198.000000
z 300.000000
rss 432.679714
Name: 61123, dtype: float64
plt.plot(y)
plt.axvline(c_opt[0], color = 'red')
plt.axvline(c_opt[1], color = 'red')
plt.axvline(c_opt[2], color = 'red')
plt.show()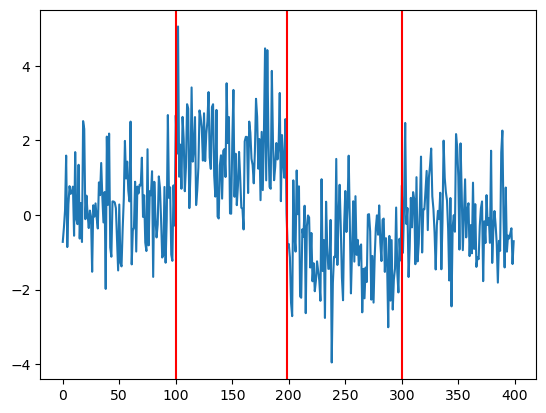
The estimates are decent.
To obtain a faster algorithm for estimating , we can try the following iterative algorithm. First we obtain by solving single change-point RSS minimization:
Then we obtain by two-change point RSS minimization with the first change-point fixed at :
Finally we determine by three-change point RSS minimization with the first change-point fixed at and the second change-point fixed at :
Here is the algorithm.
# First estimate c_1 as in the single change-point model:
def rss(c):
x = np.arange(1, n + 1)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc])
md = sm.OLS(y, X).fit()
rss = np.sum(md.resid ** 2)
return rss
allcvals = np.arange(1, n)
rssvals = np.array([rss(c) for c in allcvals])
c1_hat = allcvals[np.argmin(rssvals)]
print(c1_hat)198
# Next fix c_1 at c1_hat and estimate c_2:
def rss2(c):
x = np.arange(1, n + 1)
xc1hat = (x > c1_hat).astype(float)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc1hat, xc])
md = sm.OLS(y, X).fit()
rss = np.sum(md.resid ** 2)
return rss
allcvals = np.arange(1, n)
rss2vals = np.array([rss2(c) for c in allcvals])
c2_hat = allcvals[np.argmin(rss2vals)]
print(c2_hat)100
# Finally fix c1 at c1_hat, and c2 at c2_hat and estimate c3:
def rss3(c):
x = np.arange(1, n + 1)
xc1hat = (x > c1_hat).astype(float)
xc2hat = (x > c2_hat).astype(float)
xc = (x > c).astype(float)
X = np.column_stack([np.ones(n), xc1hat, xc2hat, xc])
md = sm.OLS(y, X).fit()
rss = np.sum(md.resid ** 2)
return rss
allcvals = np.arange(1, n)
rss3vals = np.array([rss3(c) for c in allcvals])
c3_hat = allcvals[np.argmin(rss3vals)]
print(c3_hat)300
In this example, this iterative scheme (which runs much much faster than joint grid minimization) gives the same estimates as the full grid search.