import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import statsmodels.api as smWe shall look at some standard regression terminology and also how some of the numbers shown in the regression output are actually computed.
Let us use the GDP dataset from FRED (https://
gdp = pd.read_csv('GDP-Jan2025FRED.csv')
print(gdp.head(10))
print(gdp.tail(10)) observation_date GDP
0 1947-01-01 243.164
1 1947-04-01 245.968
2 1947-07-01 249.585
3 1947-10-01 259.745
4 1948-01-01 265.742
5 1948-04-01 272.567
6 1948-07-01 279.196
7 1948-10-01 280.366
8 1949-01-01 275.034
9 1949-04-01 271.351
observation_date GDP
301 2022-04-01 25805.791
302 2022-07-01 26272.011
303 2022-10-01 26734.277
304 2023-01-01 27164.359
305 2023-04-01 27453.815
306 2023-07-01 27967.697
307 2023-10-01 28296.967
308 2024-01-01 28624.069
309 2024-04-01 29016.714
310 2024-07-01 29374.914
plt.plot(gdp['GDP'])
plt.xlabel("Time (quarterly)")
plt.ylabel("Billions of Dollars")
plt.title("Gross Domestic Product (GDP) of the United States")
plt.show()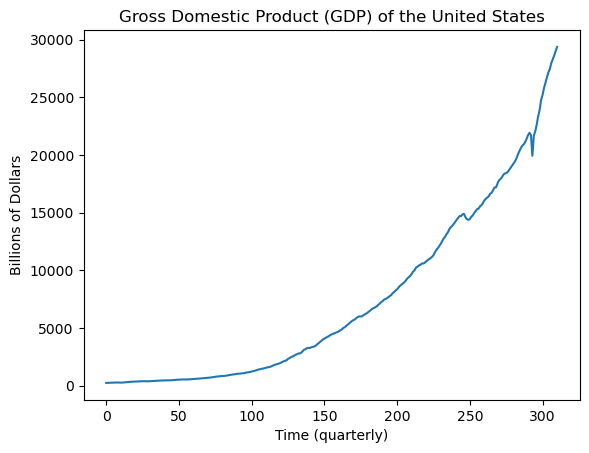
Let us fit a cubic trend model to this dataset given by:
for , where is the total number of observations in the dataset. In vector-matrix notation, this model can be equivalently represented as:
where
Observe is , is , β is , and ε is . The entries of the vector ε are known as errors, and we assume that . There are 5 parameters in this model , which we need to estimate from the data.
To implement regression, we simply need to create and and give them as input to the sm.OLS function.
The following code creates and .
y = gdp['GDP']
print(len(y))
x = np.arange(1, len(y) + 1)
x2 = x ** 2
x3 = x ** 3
X = np.column_stack([np.ones(len(y)), x, x2, x3])
#X = np.column_stack([x, x2, x3])
#X = sm.add_constant(X)
print(X[:5])311
[[ 1. 1. 1. 1.]
[ 1. 2. 4. 8.]
[ 1. 3. 9. 27.]
[ 1. 4. 16. 64.]
[ 1. 5. 25. 125.]]
Now, we simply use sm.OLS with and :
md = sm.OLS(y, X).fit()
print(md.summary()) OLS Regression Results
==============================================================================
Dep. Variable: GDP R-squared: 0.995
Model: OLS Adj. R-squared: 0.995
Method: Least Squares F-statistic: 2.000e+04
Date: Fri, 31 Jan 2025 Prob (F-statistic): 0.00
Time: 18:56:03 Log-Likelihood: -2402.2
No. Observations: 311 AIC: 4812.
Df Residuals: 307 BIC: 4827.
Df Model: 3
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 292.1841 126.498 2.310 0.022 43.271 541.097
x1 -2.5806 3.506 -0.736 0.462 -9.479 4.317
x2 0.0759 0.026 2.910 0.004 0.025 0.127
x3 0.0007 5.5e-05 12.093 0.000 0.001 0.001
==============================================================================
Omnibus: 74.600 Durbin-Watson: 0.095
Prob(Omnibus): 0.000 Jarque-Bera (JB): 746.989
Skew: 0.635 Prob(JB): 6.21e-163
Kurtosis: 10.485 Cond. No. 4.63e+07
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 4.63e+07. This might indicate that there are
strong multicollinearity or other numerical problems.
Least Squares Estimates¶
The information on the estimates along with associated uncertainties (standard errors) are given in the table near the middle of the output. Specifically, the estimates from the above table are , , , and . These numbers can also be obtained using model.params as follows:
print(md.params)const 292.184077
x1 -2.580592
x2 0.075924
x3 0.000665
dtype: float64
These estimates are known as the Least Squares Estimates (also the same as Maximum Likelihood Estimates), and they are computed via the formula:
Let us verify that this formula indeed gives the reported estimates.
XTX = np.dot(X.T, X)
#XTX = np.matmul(X.T, X)
XTX_inverse = np.linalg.inv(XTX)
XTX_inverse_XT = np.dot(XTX_inverse, X.T)
betahat = np.dot(XTX_inverse_XT, y)
print(np.column_stack([betahat, md.params]))[[ 2.92184077e+02 2.92184077e+02]
[-2.58059163e+00 -2.58059163e+00]
[ 7.59236235e-02 7.59236235e-02]
[ 6.64683542e-04 6.64683542e-04]]
In reality, sm.OLS does not use the above formula directly (even though the formula is the correct one), instead it uses some efficient linear algebra methods to solve the linear equations .
Fitted Values¶
The next quantity to understand are the fitted values. These are given by model.fittedvalues and they are obtained via the simple formula:
The entries of the vector are known as the fitted values. Let us check the correctness of this formula.
fvals = np.dot(X, betahat)
print(np.column_stack([fvals[:10], md.fittedvalues[:10]]))[[289.68007406 289.68007406]
[287.33190609 287.33190609]
[285.14356157 285.14356157]
[283.1190286 283.1190286 ]
[281.26229528 281.26229529]
[279.57734972 279.57734972]
[278.06818001 278.06818001]
[276.73877426 276.73877426]
[275.59312056 275.59312056]
[274.63520702 274.63520702]]
To assess how well the regression model fits the data, we plot the fitted values along with the original data.
plt.plot(y, label = "Data")
plt.plot(md.fittedvalues, label = "Fitted values")
plt.legend()
plt.xlabel("Time (quarterly)")
plt.ylabel("Billions of Dollars")
plt.title("Gross Domestic Product (GDP) of the United States")
plt.show()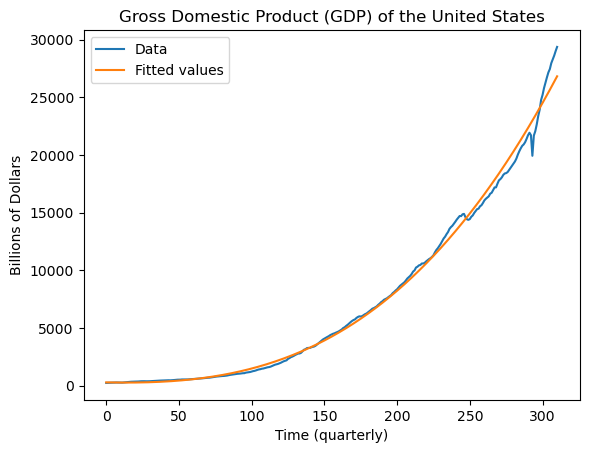
Residuals¶
The residuals simply are the differences between the observed data and the fitted values. We use the notation for the vector of residuals:
The residual is simply the entry of .
residuals = y - fvals
print(np.column_stack([md.resid[:10], residuals[:10]]))[[-46.51607406 -46.51607406]
[-41.36390609 -41.36390609]
[-35.55856157 -35.55856157]
[-23.3740286 -23.3740286 ]
[-15.52029529 -15.52029528]
[ -7.01034972 -7.01034972]
[ 1.12781999 1.12781999]
[ 3.62722574 3.62722574]
[ -0.55912056 -0.55912056]
[ -3.28420702 -3.28420702]]
A plot of the residuals against time is an important diagnostic tool which can tell us about the effectiveness of the fitted model.
plt.plot(md.resid)
plt.xlabel("Time (quarterly)")
plt.ylabel("Billions of Dollars")
plt.title("Residuals")
plt.show()
From this plot, we see that there are some very large residuals indicating the presence of time points where the model predictions are quite far off from the observed values. This could be because of outliers or some systematic features that the model is missing. Also the residual plot looks quite smooth which indicates that there is some correlation between nearby residuals. This can be better visualized via the acf (autocorrelation function) plot (sometimes known as the correlogram) of the residuals.
print(gdp[:10])
gdp['observation_date'][np.argmin(residuals)] observation_date GDP
0 1947-01-01 243.164
1 1947-04-01 245.968
2 1947-07-01 249.585
3 1947-10-01 259.745
4 1948-01-01 265.742
5 1948-04-01 272.567
6 1948-07-01 279.196
7 1948-10-01 280.366
8 1949-01-01 275.034
9 1949-04-01 271.351
'2020-04-01'ACF Plot¶
Given a time series dataset and a lag h, define
for . Here is simply the mean of . The acf plot graphs on the x-axis and on the y-axis. The quantity is known as the sample autocorrelation of the data at lag (acf stands for “Autocorrelation Function”). When is large and is small, approximates the sample correlation in the bivariate dataset . The actual formula for the sample correlation in the bivariate dataset is:
If we now use the simple approximations and and replace the sums in the denominator to range over all (as opposed to ), we get the formulat for . These approximations are reasonable when is large and is small.
The ACF plot tells us about the size of the correlations between the successive values of given time series. Note that is always equal to 1. So we are really looking at the size of for .
Here is the ACF plot for the residuals of the regression fitted to the GDP data.
from statsmodels.graphics.tsaplots import plot_acf
plot_acf(md.resid, lags = 50)
plt.xlabel("Lag")
plt.ylabel("Autocorrelation")
plt.title("Autocorrelation Function of Residuals")
plt.show()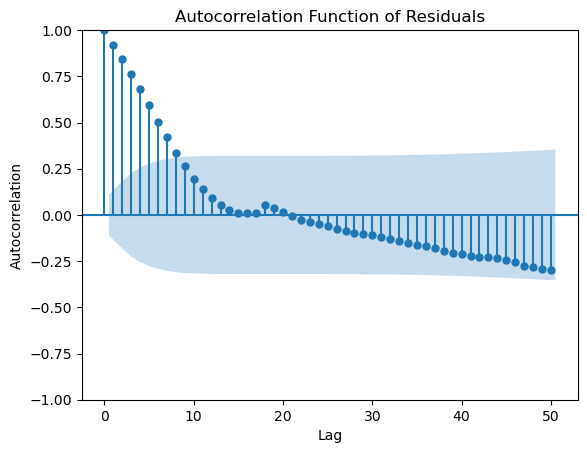
This plot reveals that the residuals have significant autocorrelations. Note the presence of the blue shaded region that is automatically supplied by the plot. This is supposed to help us assess the size of the autocorrelations. The idea is that even if the data is i.i.d so that there is are no autocorrelations, by randomness, some of the computed sample autocorrelations will be nonzero. The typical size of these null autocorrelations is indicated by the blue shaded region. The implication is that we should only consider the size of an autocorrelation as significantly different from zero if it sticks out of the blue regions.
iidz = np.random.normal(size = 400)
plot_acf(iidz, lags = 50)
plt.xlabel("Lag")
plt.ylabel("Autocorrelation")
plt.title("Sample autocorrelations of i.i.d noise")
plt.show()
#note that the acf value at h = 0 is always 1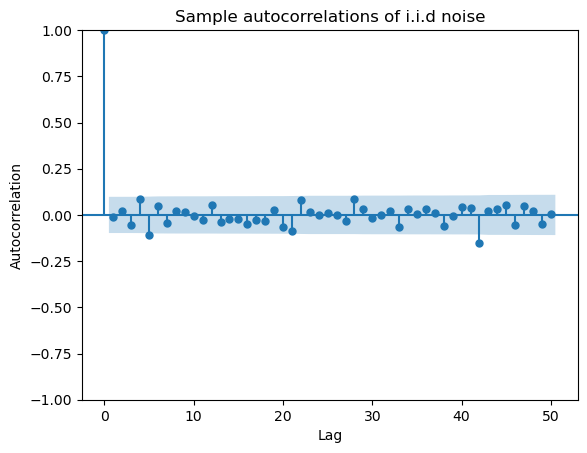
Let us get back to the ACF plot of the residuals from the model fit to the GDP data. There are significant autocorrelations at small lags (especially ). The implication of this is the following. Suppose we want to predict the GDP for the future quarter immediately following the last data observation. We can use the predicted value given by the model. But the last residual value is about 2548, which is quite larger than zero. Because of significant positive autocorrelation at lag 1, we would expect the next residual value to be quite positive as well. This means that we should adjust the predicted value given by the model upwards by about 2548 for a better forecast. This makes sense from the plot of the data and fitted values as well. We shall study such procedures later in the course.
#the value of the last residual
md.resid[len(y) - 1]np.float64(2548.0506349269126)Residual Sum of Squares (RSS) and Residual df¶
Two other commonly used terms (in connection with residuals) are the Residual Sum of Squares (RSS) and the Residual Degrees of Freedom. The RSS is simply the sum of the squares of the residuals:
RSS is simply equal to the smallest possible of the sum of squares criterion (recall from lecture that ).
The vector of residuals has the following important property: . This can be proved as follows:
means that the dot product between every column of and equals 0. In our regression model for GDP, is equivalent to:
Even though there are residuals , they have to satisfy the above four equality constraints. Therefore, the effective number of ‘free’ residuals is . Thus, the residual degrees of freedom equals .
More generally, the residual df equals the number of observations () minus the number of columns in the matrix.
Estimate of σ: the residual standard error¶
The estimate of σ is given by:
This quantity is sometimes called the Residual Standard Error. The value of the Residual Standard Error can be used to assess the size of the residuals (residuals much larger than say twice the Residual Standard Error indicate points where the model fits particularly poorly).
rss = np.sum(md.resid ** 2)
rse = np.sqrt(rss/(len(y) - 4))
print(rse)550.9932711741246
This intuitively means that residuals with magnitude above 1100 are points where the model fits particularly poorly. From a look at the plot of the residuals against time, there are many residuals which are this large in magnitude.
Standard Errors of the coefficient estimates¶
The standard errors corresponding to are the square roots of the diagonal entries of . They are given by md.bse as verified below.
sebetahat_squared = (rse ** 2) * (np.diag(XTX_inverse))
sebetahat = np.sqrt(sebetahat_squared)
print(np.column_stack([md.bse, sebetahat]))[[1.26498064e+02 1.26498064e+02]
[3.50560340e+00 3.50560340e+00]
[2.60867093e-02 2.60867093e-02]
[5.49661580e-05 5.49661580e-05]]
t-statistic and confidence intervals for the coefficients¶
The -statistic for each coefficient is simply the estimate divided by the standard error. The confidence interval for is given by:
where is the point beyond the -distribution with degrees of freedom gives probability mass . The above formula is the result of:
The above statement can also be interpreted in a frequentist sense (then the conditional on data will be removed and the randomness will be over the data with fixed ).
from scipy.stats import t
alpha = 0.05
qt = t.ppf(1 - (alpha/2), len(y) - 4) # area to the right equaling alpha/2 is the same as area to the left equaling 1-alpha/2
print(qt)
cilower = md.params - md.bse * qt
ciupper = md.params + md.bse * qt
print(np.column_stack([cilower, ciupper]))
print(md.summary())1.9677212881552213
[[ 4.32711446e+01 5.41097010e+02]
[-9.47864205e+00 4.31745880e+00]
[ 2.45922503e-02 1.27254997e-01]
[ 5.56525463e-04 7.72841622e-04]]
OLS Regression Results
==============================================================================
Dep. Variable: GDP R-squared: 0.995
Model: OLS Adj. R-squared: 0.995
Method: Least Squares F-statistic: 2.000e+04
Date: Fri, 31 Jan 2025 Prob (F-statistic): 0.00
Time: 18:56:03 Log-Likelihood: -2402.2
No. Observations: 311 AIC: 4812.
Df Residuals: 307 BIC: 4827.
Df Model: 3
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 292.1841 126.498 2.310 0.022 43.271 541.097
x1 -2.5806 3.506 -0.736 0.462 -9.479 4.317
x2 0.0759 0.026 2.910 0.004 0.025 0.127
x3 0.0007 5.5e-05 12.093 0.000 0.001 0.001
==============================================================================
Omnibus: 74.600 Durbin-Watson: 0.095
Prob(Omnibus): 0.000 Jarque-Bera (JB): 746.989
Skew: 0.635 Prob(JB): 6.21e-163
Kurtosis: 10.485 Cond. No. 4.63e+07
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The condition number is large, 4.63e+07. This might indicate that there are
strong multicollinearity or other numerical problems.
Visualizing Uncertainty¶
It was showed in Lecture 4 that the posterior distribution of β (this is the vector of coefficients) is given by
From the definition of the -distribution above, we also know that the distribution above coincides with the distribution of
where and are independent. Using this, we can generate vectors β from their posterior distribution and then plot the regression curves corresponding to the different generated β vectors. This will give us an idea of the uncertainty underlying the coefficients.
N = 200 # this the number of posterior samples that we shall draw
Sigma_mat = (rse ** 2) * (XTX_inverse)
#print(Sigma_mat)
rng = np.random.default_rng()
chi_samples = rng.chisquare(df = len(y) - 4, size = N)
chi_samples = chi_samples.reshape((-1, 1))
norm_samples = rng.multivariate_normal(mean = np.zeros(4), cov = Sigma_mat, size = N)
#post_samples = np.tile(betahat, (N, 1)) + norm_samples / np.sqrt(chi_samples / (len(y) - 4))
post_samples = betahat + norm_samples / np.sqrt(chi_samples / (len(y) - 4))
#print(post_samples)
plt.figure(figsize = (10, 6))
plt.plot(y, linewidth = 1, color = 'black')
for k in range(N):
fvalsnew = np.dot(X, post_samples[k])
plt.plot(fvalsnew, color = 'red', linewidth = 1)
plt.xlabel('Time (quarterly)')
plt.ylabel('Billions of dollars')
plt.title('GDP data with Fitted values along with uncertainty')
plt.show()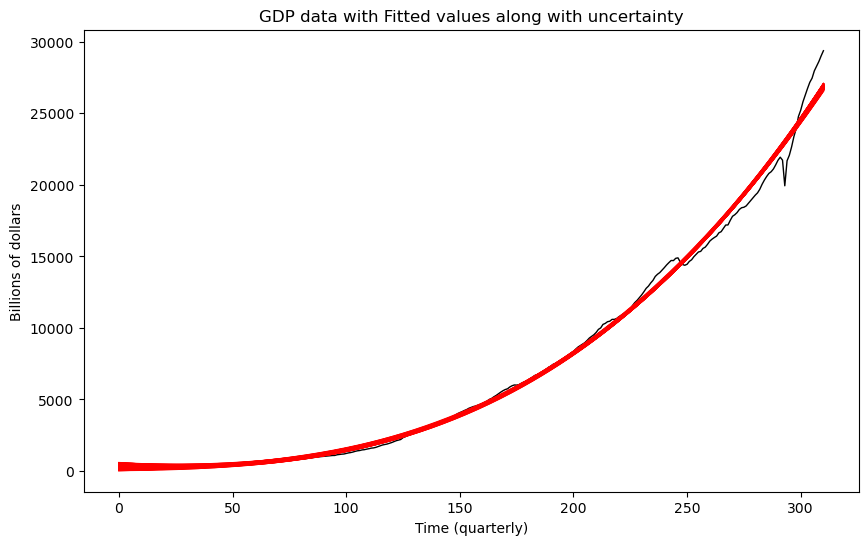
Even though regression curves have been plotted, they are still fairly clustered together. This suggests that the uncertainty in these estimates is fairly small.
Other comments¶
When analyzing such a dataset, it is also common to work with logs instead of the raw GDP numbers directly i.e., take . Another common approach is to work with difference of logs (this is interpreted as the GDP growth rate): . If you want, please show the fitted regression equation for one of these two modified responses. For prediction purposes, note that if you predict a future value of the difference of the logs, then this predicted value can be used to also obtain a prediction for the original data. Please show how this may be done if possible.